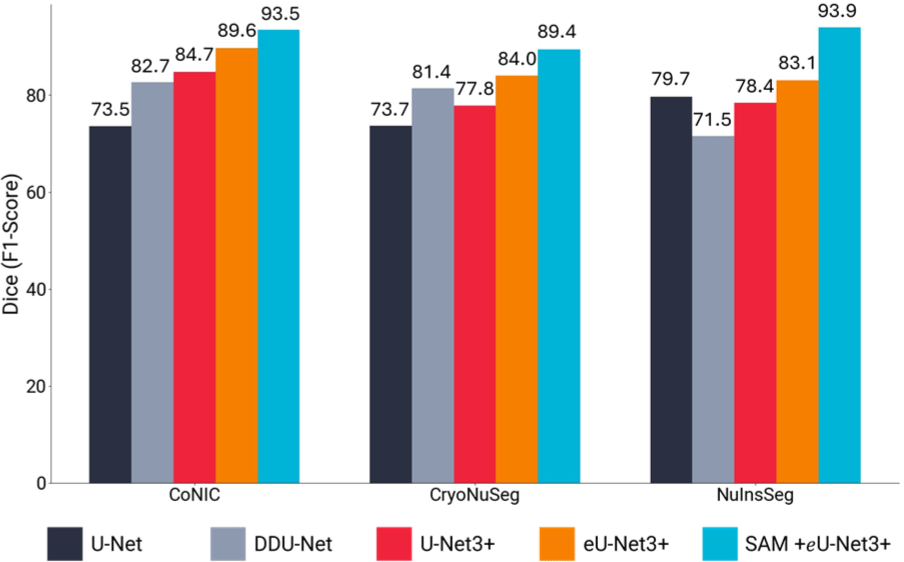I. INTRODUCTION
In digital pathology, accurate segmentation of nuclei in histological images is vital for cancer diagnosis and prognosis [1]. Segmentation of nuclei enables detailed examination of cellular structures and behaviors, such as analyzing cell cycles, mutations, and the morphology of cancerous tissues [2]. These analyses are essential for identifying cancer types, evaluating severity, and guiding treatment plans [3]. Tissue biopsy-based diagnostics relies heavily on this type of segmentation and remains the gold standard for cancer detection worldwide [4]. Given the volume and complexity of biopsy samples, automated and accurate segmentation methods have become critical to aid pathologists and streamline diagnostic workflows.
Nuclei segmentation is a fundamental step in quantitative histopathology that enables the analysis of cellular morphology, spatial distribution, and tissue architecture [4]. Accurate segmentation is crucial for diagnosing various diseases, including cancer, where nuclear features are key indicators of malignancy [5]. Traditional manual annotation is time-consuming and subject to inter-observer variability [6], highlighting the need for automated solutions.
Task-specific models, such as the U-Net family of architectures, have emerged as the benchmark for medical image segmentation [5]. U-Net [6] variants are effective in capturing the fine-grained details that are necessary for high-precision cell segmentation. They are effective due to their encoder-decoder structure and skip connections which help preserve contextual information. Despite their success, task-specific models typically require large amounts of labeled training data and often struggle to generalize across different staining variations and imaging modalities that are commonly encountered in histopathological datasets [7]. Moreover, these models tend to focus on localized features and may not fully capture the broader context of tissue structures, which is crucial for understanding complex cell and tissue interactions [8-9]. Moreover, challenges such as heterogeneity in staining, presence of artifacts, and variability in nuclear morphology necessitate more robust approaches.
To address the above limitations, recent research has explored the potential of foundation models, such as the Segment Anything Model (SAM) [10], which are trained on vast and diverse datasets. Foundation models excel in capturing global contextual features and generalize well across various visual domains. However, when applied to histology images, foundational models often lack the pixel-level precision needed for accurate pathological analysis, especially in segmenting small, densely packed nuclei [7]. Our study aims to address these challenges by integrating global contextual information from foundation models with the precision of task-specific models.
In this paper, we propose a novel approach that integrates the strengths of task-specific and foundation models to enhance nuclei segmentation in histology images. We enhance the U-Net3+ architecture [11] by introducing an adaptive feature selection mechanism, which we call, eU-Net3+. Additionally, we propose an Enhanced Fusion Block (EFB), which dynamically fuses the global contextual knowledge from foundation models with the detailed local representations from task-specific models using cross-attention and gated squeeze-and-excitation techniques [12]. Our proposed framework enables the model to leverage both global context and local precision, addressing the challenges posed by complex histological images.
Our approach demonstrates significant improvements in segmentation performance, achieving a 12% and 17.22% increase in Dice score and mIoU, respectively, on the CryoNuSeg dataset [13], a 15.55% and 16.77% increase on the NuInsSeg dataset [14], and a 9% improvement on both metrics for the CoNIC dataset [15]. By effectively merging task-specific models with foundation models, we set a new standard for state-of-the-art nuclei segmentation in digital pathology. The main contributions of this paper are as follows:
-
Integration of Task-Specific and Foundation Models: We propose a framework that effectively combines the fine-grained feature extraction capabilities of task-specific models with the global contextual understanding of foundation models. Our Enhanced Fusion Block (EFB) dynamically fuses local and global features through cross-attention and gated squeeze-and-excitation techniques.
-
Adaptive Feature Selection using GLUs: We introduce an adaptive feature selection mechanism using Gated Linear Units (GLUs) within the U-Net3+ architecture to create eU-Net3+, which enhances local feature extraction and improves segmentation accuracy.
-
State-of-the-Art Nuclei SegmentationResults: By setting new benchmarks on the CryoNuSeg, NuInsSeg, and CoNIC datasets, our work advances the field of nuclei segmentation in digital pathology.
II. RELATED WORKS
Nuclei segmentation in histopathological images has witnessed significant advances with the advent of deep learning, particularly convolutional neural networks (CNNs) [6]. Among CNN-based architectures, U-Net has become a seminal model for medical image segmentation due to its symmetrical encoder-decoder structure and the use of skip connections, which preserve spatial details while extracting higher-level features [16]. Variants of U-Net [16-19] have demonstrated remarkable success in various biomedical tasks, including nuclei segmentation, by improving multi-scale feature extraction and incorporating attention mechanisms.
Recent approaches have focused on hybrid models that combine U-Net with other architectures to leverage the strengths of both [20]. These hybrid approaches address the limitations of U-Net in handling complex visual features like overlapping nuclei, irregular shapes, and varying sizes. For instance, ASPPU-Net [21] integrates Atrous Spatial Pyramid Pooling (ASPP) with U-Net, enhancing its ability to capture multi-scale contextual information. Similarly, Hover-Net [22] extends U-Net with residual connections and dense blocks to improve feature reuse and boundary precision, particularly in dense cell regions. Another notable advancement is the Sharp U-Net models [23], which aim to increase performance by minimizing low-frequency noise introduced during down-sampling and up-sampling layers. Attention mechanisms have also become a popular enhancement to U-Net variants. DEAU [24] introduces an Attention Encoding Path (AEP) that runs parallel to the U-Net’s traditional encoding path, refining feature extraction by using attention maps that prioritize diagnostically significant regions. Similarly, methods incorporating self-attention or cross-attention layers have shown improved nuclei detection and segmentation accuracy by capturing long-range dependencies and suppressing irrelevant background information. DDU-Net [25] leverages dual decoders to handle both nuclear and cytoplasmic regions, enhancing segmentation performance on histopathological images where overlapping structures are prevalent. U-Net3+ [11], on the other hand, focuses on incorporating full-scale skip connections and deep supervision to better fuse multi-scale features. To benchmark our proposed model, DDU-Net and U-Net3+ were selected as baseline models as they previously obtained state-of-the-art results. Both architectures are well-regarded for their effectiveness in medical image segmentation. While U-Net3+ excels in fusing full-scale feature maps and providing deep supervision, DDU-Net’s dual-decoder design allows it to effectively differentiate between different cellular structures, making these models strong candidates for comparison.
Additionally, some studies have employed wavelet-based channel attention modules to capture more global context within U-Net-based models [26]. By decomposing feature maps into different frequency components using wavelet transforms can help these methods effectively focus on salient features at various scales and enhance segmentation performance by integrating both local and global information. Post-processing techniques have also been explored to improve segmentation results. Studies like [27] have used morphological operations to refine segmentation masks. Semi-supervised [28-30] and unsupervised learning approaches [31-34] have also been investigated for nuclei segmentation, aiming to reduce the reliance on large amounts of annotated data. Methods such as [35,36] utilize generative adversarial networks (GANs) or self-supervised learning techniques to learn representations from unlabeled data. However, these methods often struggle to achieve high performance due to the complexity and variability of histopathological images, and the lack of explicit guidance from labeled examples limits their effectiveness compared to fully supervised approaches.
Another trend is the integration of foundation models such as Vision Transformers (ViTs) [37] that are pre-trained on vast and diverse datasets. Foundation models offer strong generalization across various domains by learning rich, global contextual features, as demonstrated by the Segment Anything Model (SAM) [10]. While these models excel at capturing global context, their application in medical image segmentation, particularly nuclei segmentation, requires finetuning or parameter-based optimizations. SAM is trained on natural images that lacks the fine-grained detail necessary for precise nuclei boundary detection, making it necessary to finetune or adapt it to task-specific demand like nuclei segmentation. There have been recent developments that include MedSAM [38] which is specifically trained on medical images. But even then, it is far from reaching the performance level of task-specific models semantic segmentation models.
Our work builds on these developments by proposing a novel hybrid model that combines the global context awareness of SAM with the fine-grained feature extraction capabilities of U-Net. By incorporating GLU and GFB, our approach dynamically and effectively fuses local and global features, allowing the model to address the challenges of nuclei segmentation in complex histopathological images. While many previous models focus on either task-specific or general foundation models, our approach effectively combines both to achieve state-of-the-art performance, as demonstrated by our significant improvements in Dice scores across multiple datasets.
III. METHODS
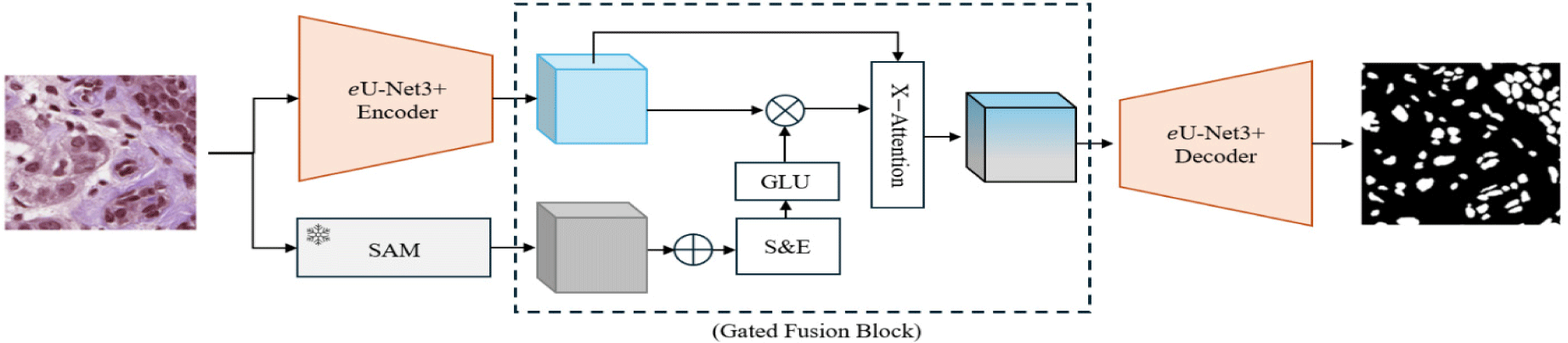
The overall pipeline of the proposed approach is depicted in Fig. 1. Our methodology focuses on enhancing task-specific nuclei segmentation by leveraging the combined strengths of a task-specific model and a foundation model. The task-specific model is an enhanced version of U-Net3+ (referred to as eU-Net3+), optimized for fine-grained feature extraction. In parallel, a pre-trained Segment Anything Model (SAM) provides global contextual information. These local and global features are fused through our proposed Enhanced Fusion Block (EFB), which combines Gated Linear Units (GLUs) in a squeeze-and-excitation mechanism followed by a cross-attention block. This ensures effective integration of both global and local representations for enhanced segmentation performance.
The U-Net architecture has emerged as a fundamental framework in medical image segmentation due to its unique ability to balance both high-level semantic information and low-level spatial details [20]. Over time, numerous variants of the U-Net architecture have been proposed to address specific challenges in medical image segmentation. Most variations of U-Net introduce enhancements to tackle particular issues such as multi-scale feature extraction and deeper supervision. From among the U-Net based models, U-Net3+ introduces full scale skip connections and allows features from all levels of the encoder to be directly connected to the corresponding layers in the decoder. This deep supervision improves gradient flow during training and facilitates better feature learning by incorporating multiple output layers. While U-Net3+ provides a strong foundation for segmenting histopathological images, traditional activation functions such as ReLU [39] apply uniform transformations to all features and overlook the nuances in densely packed or morphologically diverse regions. To address this limitation, we enhance U-Net3+ by GLUs, allowing for adaptive feature selection.
GLUs split the input into two streams: one undergoes a linear transformation, while the other passes through a sigmoid activation function. The sigmoid activation serves as a gate, modulating the flow of information based on the relevance of each feature to the segmentation task. This mechanism allows the network to selectively focus on diagnostically significant regions, such as densely packed nuclei, while ignoring less relevant features. The GLU operation is defined as:
where, ⊙ denotes element-wise multiplication, Wa,ba represent the weights and biases for gating mechanism, and Wl,bl represent the weights and biases for the linear transformation. This enhancement ensures that the task-specific model not only captures multi-scale features but also fine-tunes its focus towards the most significant regions in the image.
The Segment Anything Model (SAM) [10] is used as a foundation model. SAM is pre-trained on a vast dataset comprising over 11 million images and one billion masks. Although SAM is not specialized for medical images, its robust ability to capture high-order global context across diverse visual domains makes it a valuable asset for guiding nuclei segmentation. SAM’s ability to identify global contextual relationships aids the task-specific model in understanding broader tissue structures, which is particularly helpful when analyzing complex histological images.
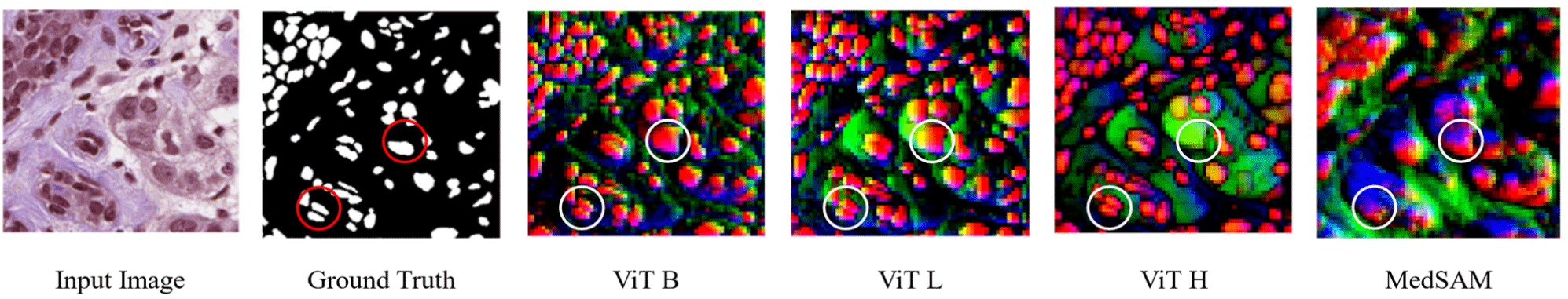
In our proposed framework, we use SAM’s Base model checkpoint (91M parameters) – ViT B, as it provides a more abstract and ambiguous representation of the input image, which complements the detailed focus of eU-Net3+ as can be seen in Fig. 2. This ambiguity allows the task-specific model to fine-tune its decisions, particularly in differentiating nuclei from surrounding regions. Visualizing SAM’s encodings through Principal Component Analysis (PCA), we observe that simpler SAM representations produced by the base ViT enable a better fusion with the local features captured by eU-Net3+.
A key challenge when combining feature representations from task-specific and foundation models is the potential for conflicting or redundant information. Simple concatenation of features often results in suboptimal performance due to this misalignment [40]. To address this, we introduce Enhanced Fusion Block (EFB) to effectively integrate the global context with local task-specific features.
The EFB consists of three main components: a Gated Squeeze-and-Excitation block, GLU block and a Cross-Attention Block. The squeeze-and-excitation block starts by performing adaptive average pooling on the concatenated features from SAM and eU-Net3+. This compresses spatial dimensions to focus on global information from each channel. The gated mechanism, implemented using GLUs, selectively allows important features to flow through, emphasizing only the most relevant global and local features. The cross-attention block operates on the gated features by treating them as queries, keys, and values in a standard attention operation. This mechanism enhances the model’s ability to highlight important features, suppress irrelevant information, and increase contextual awareness.
IV. EXPERIMENTS AND RESULTS
We used three publicly available histopathological datasets that contained variability in staining techniques, and tissue morphology.
-
CryoNuSeg [13]: This dataset consists of 30 high-resolution images obtained using cryo-sectioning techniques. It is known for its variability in nuclear morphology and presents challenges related to segmentation accuracy due to its complex staining and tissue structures.
-
NuInsSeg [14]: Comprising 665 image patches, NuInsSeg is a challenging dataset characterized by diverse tissue types and staining methods. The dataset includes densely packed nuclei and complex tissue structures, making it an ideal test bed for assessing segmentation performance on intricate regions.
-
CoNIC [15]: The CoNIC dataset includes 4,981 images and presents one of the largest and most complex datasets used for nuclei segmentation. It contains multiple tissue types and wide variations in nuclear shapes, providing a robust benchmark for evaluating generalization across diverse histological samples.
Our experiments were conducted using the PyTorch framework on a system equipped with an NVIDIA RTX A6000 GPU. To ensure consistency and standardization, all images were resized to a resolution of 256×256 pixels. The training process spanned 50 epochs, with an initial learning rate of 1×10−4 and a batch size of 16. We employed the Adam optimizer [41], which is well-suited for segmentation tasks, and incorporated a dropout rate of 0.3 to prevent overfitting.
All the images were preprocessed by using stain normalization as described in [42] and then augmentations were performed. We applied a combination of photometric and geometric augmentations as described in [43]. Geometric augmentations such as rotations, scaling, flips were performed to obtain samples from different perspectives and scales. Additionally, elastic deformations that mimic the natural deformations in biological tissues were performed that helped in enhancing the model’s ability to handle non-rigid transformations and complex variations. Photometric augmentations like Gamma and intensity level transformations along with contrast limited adaptive histogram equalization were performed. These augmentations aimed to increase the diversity of the training set and help the model generalize better to unseen data.
For the loss function, we used a weighted combination of Dice loss [43] and focal loss [45], each contributing equally to the overall loss function. Dice loss was chosen for its effectiveness in handling imbalanced datasets, particularly in scenarios where nuclei occupy a small portion of the image. Focal loss further helps by focusing on hard-to-segment regions, ensuring that rare and challenging nuclei instances are not overlooked during training.
To evaluate the segmentation performance, we used the Dice coefficient and mean Intersection over Union (mIoU), both of which are standard metrics in medical image segmentation tasks. These metrics provide a robust evaluation of the overlap between the predicted segmentation masks and the ground truth, with higher values indicating better segmentation quality.
The experimental results are summarized in Table 1. It demonstrates that integrating SAM’s global contextual features significantly improves the performance of the task-specific eU-Net3+ model across all datasets.
-
CryoNuSeg: Our model achieved a 12% increase in Dice score and a 17.22% increase in mIoU compared to baseline models. The inclusion of SAM helped mitigate the effects of freezing artifacts by providing additional context to differentiate nuclei from artifacts.
-
NuInsSeg: We observed over 15% improvement in Dice score and mIoU. The model effectively handled densely packed and overlapping nuclei by leveraging global contextual information from SAM, aiding in distinguishing individual nuclei in crowded regions.
-
CoNIC: The model showed a 9% improvement in both Dice score and mIoU, demonstrating its ability to generalize across diverse tissue types despite significant variability in nuclear appearance.
These performance differences across datasets can be attributed to the unique characteristics and challenges presented by each dataset. Our proposed methodology even outperforms the recently released AWGUNET [26] that uses wavelet for guiding task-specific U-Net model.
The choice of activation function has a substantial impact on segmentation performance and is detailed in Table 2. The inclusion of GLUs as activation function in the eU-Net3+ architecture has significantly outperformed other commonly used activation functions, such as ReLU, LeakyReLU [46], and Swish [47], across all datasets.
| Activation functions | CryoNuSeg | NuInsSeg | CoNIC |
|---|---|---|---|
| ReLU | 0.778 | 0.7844 | 0.8474 |
| LeakyReLU | 0.7931 | 0.8012 | 0.8567 |
| Swish | 0.7583 | 0.7721 | 0.8439 |
| GLU | 0.8401 | 0.8307 | 0.8966 |
The GLU-enhanced model achieved a Dice score of 0.8401 on CryoNuSeg, 0.8307 on NuInsSeg, and 0.8966 on CoNIC, surpassing ReLU, LeakyReLU, and Swish by notable margins. This demonstrates the superiority of GLU in enabling selective feature activation, allowing the model to focus on the most relevant regions of the image for precise segmentation, especially in complex histological samples. The gating mechanism of GLU provides an adaptive feature selection that dynamically activates diagnostically important features, leading to more refined segmentation results.
The effectiveness of the proposed Gated Fusion Block (GFB) is evident from the performance gains observed. As shown in Table 3, incorporating the GFB into the model resulted in marked improvements across all datasets. The eU-Net3+ model with GFB achieved higher Dice scores and mIoU values compared to the version without GFB and shows the importance of effective feature fusion in enhancing segmentation accuracy. The GFB effectively addressed the challenge of fusing local and global features by employing GLUs for selective gating and a cross-attention mechanism for feature alignment. This allowed the model to dynamically balance fine-grained local features and broader contextual insights, leading to more accurate segmentations.
| Model | CryoN uSeg | NuInsSeg | CoNIC |
|---|---|---|---|
| eU-Net3+ | 0.8401 | 0.8307 | 0.8966 |
| eU-Net3+ w/o GFB | 0.8235 | 0.8146 | 0.8918 |
| eU-Net3+w/GFB | 0.8942 | 0.9399 | 0.9351 |
In addition, we evaluated different variants of the SAM model. As shown in Table 4, the ViT-B (Base) model consistently outperformed the larger ViT-L and ViT-H models. The ViT-B model provided an optimal level of global context without overwhelming the task-specific model with excessive detail, allowing for better integration and improved segmentation results.
| Frozen models | CryoNuSeg | NuInsSeg | CoNIC |
|---|---|---|---|
| ViT - B | 0.8942 | 0.9399 | 0.9351 |
| ViT - L | 0.8745 | 0.8918 | 0.9072 |
| ViT - H | 0.8731 | 0.8867 | 0.8917 |
The primary reason for the superior performance of the ViT-B (Base) model can be attributed to its ability to maintain a more balanced level of ambiguity in its representations. Although the ViT-H (Huge) model, with its 636M parameters, provides more detailed and nuanced representations of the input images, can lead to the loss of necessary ambiguity in certain regions of the histological images, as illustrated in Fig. 2. The ViT-B model, with 91M parameters, provided a more optimal level of ambiguity in global context which allowed the eU-Net3+ task-specific model to make finer decisions in ambiguous regions, such as distinguishing between nuclei and non-nuclei areas. By allowing the task-specific model to handle these more nuanced decisions, rather than relying entirely on SAM’s global representation, the proposed model was able to achieve more accurate segmentation results.
Fig. 3 visually depicts the performance of various models across datasets. The violin plots in Fig. 4 provide further insight into the distribution of Dice scores for models with and without SAM integration. Mean Dice Scores (dotted lines) show a clear improvement with SAM integration across all datasets.
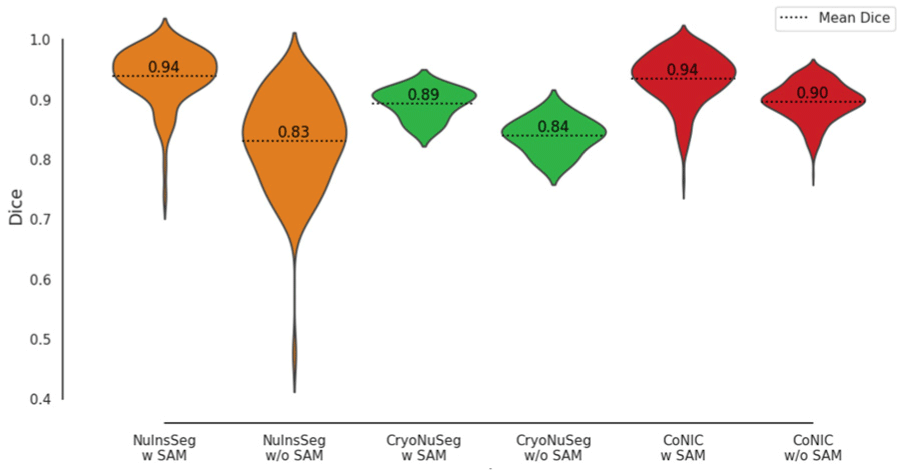
The violin plot in Fig. 4 also illustrates the variance in Dice scores, showing that SAM-guided models produce more consistent and reliable results, with less variance in their predictions.
Fig. 5 shows the qualitative assessment of the proposed model. The circles areas in the image highlights some of the regions where inclusion of SAM in eU-Net3+ performed better than using only the task-specific model. The SAM guided predictions generally show more precise and accurate segmentation, further confirming the effects of the proposed method of adding global context from SAM to improve the segmentation accuracy of the task-specific model.
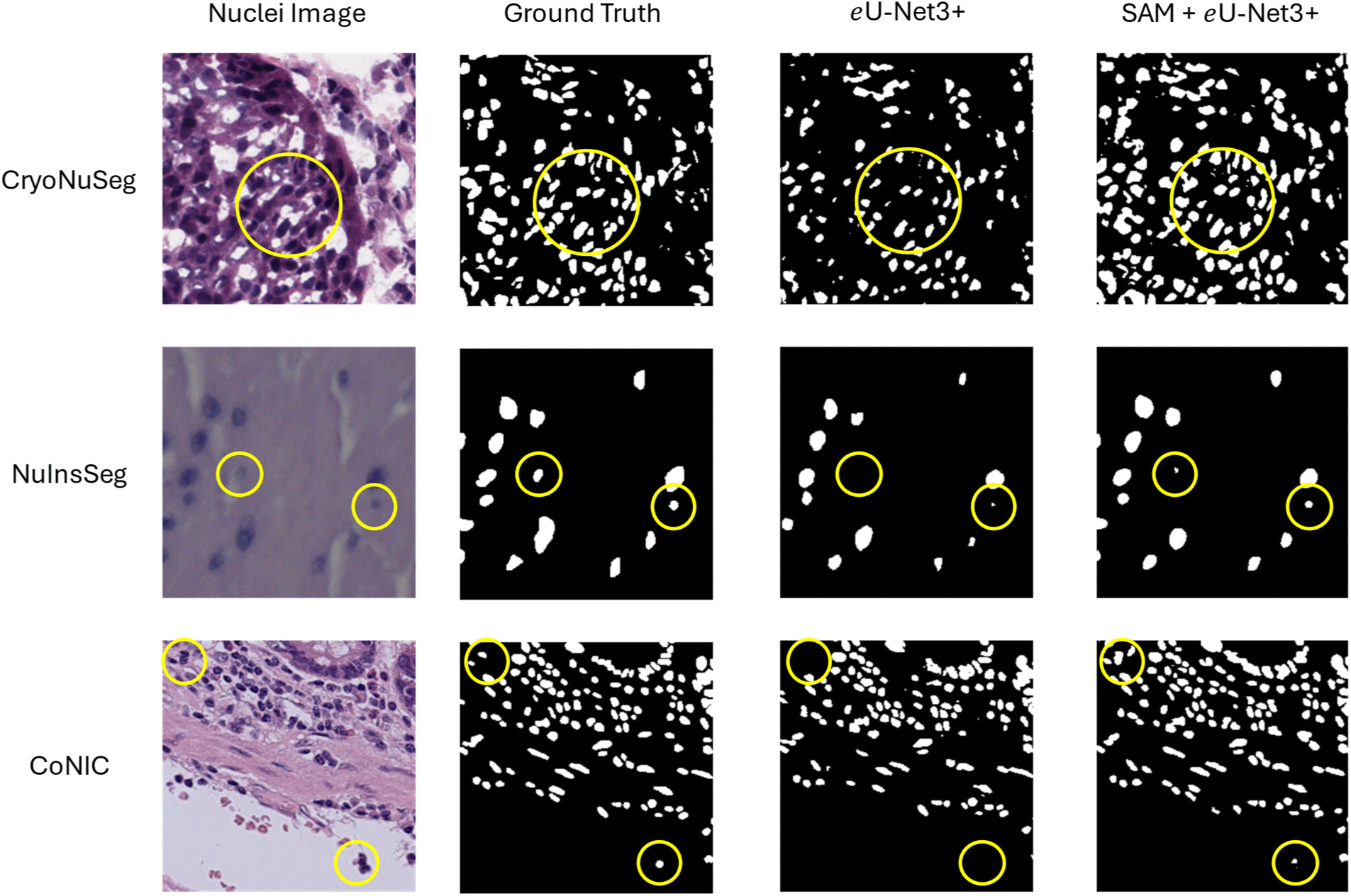
There are performance differences between three different datasets as the images in the datasets contain varied properties. In the data collection of histopathology images various techniques are used for creating the whole slide images. For instance, CryoNuSeg contains cryo-sectioned images that may include freezing artifacts affecting image clarity; NuInsSeg features densely packed nuclei from diverse tissues with varying staining techniques, making segmentation challenging; and CoNIC encompasses images from multiple organs with different staining protocols, resulting in significant variability in nuclear appearance. These differences contribute to the varied performance of our model across the datasets.
V. CONCLUSION
In this paper, we introduced a novel method for nuclei segmentation in histopathological images. We integrated task-specific models with foundation models to enhance performance. Specifically, we improved the U-Net3+ architecture by incorporating GLUs for adaptive feature selection. We also proposed GFB that dynamically fuses local (from eU-Net3+) and global representations (from SAM) using cross-attention and gated squeeze-and-excitation techniques. This fusion of global and local features enabled our model to tackle challenges such as varying tissue structures, complex staining techniques, and densely packed nuclei. Our experiments on three challenging histopathological datasets—CryoNuSeg, NuInsSeg, and CoNIC—demon-strated significant improvements in segmentation accuracy. Incorporating SAM’s global context led to Dice score improvements of up to 12% on CryoNuSeg, 15.55% on NuInsSeg, and 9% on CoNIC compared to baseline models. Our analysis revealed that the ViT-B variant of SAM outperformed larger ViT models. It provided an optimal balance between capturing global context and computational efficiency. Visual assessments confirmed that SAM-guided models produced more accurate and reliable segmentations with reduced variance in performance. By effectively merging task-specific models with foundation models, our approach obtains the state-of-the-art results in nuclei segmentation. It also presents a flexible framework for integrating local and global representations in medical imaging. Future research can explore further optimizations of the fusion mechanisms. This includes experimenting with different attention strategies or adaptive weighting between local and global features.